A new study by the Psychiatric Genomics Consortium has identified 44 genetic loci that are linked to major depression in humans. The research was published in the journal Nature Genetics.
This is the largest study to date looking at genetic risk factors for depression, a condition that affects more than 300 million people around the world.
The reason for the significant amount of media attention this article has received is predominantly because previous studies have only managed to highlight a few genetic associations for depression. Indeed, 30 of the 44 loci identified in this study are new/novel associations that have not previously been associated with depression.
Professor Cathryn Lewis, a co-author on the paper said: “Depression is an incredibly common disorder that affects millions of people in the UK. This study has shed a bright light on the genetic basis of depression, but it is only the first step. We need further research to uncover more of the genetic underpinnings, and to understand how genetics and environmental stressors work together to increase risk of depression.”
A genome-wide association study (GWAS) was conducted; in which genetic variation in different individuals was tested on a genome-wide scale to see which loci are associated with particular traits. To do this the researchers collated data on half a million people, almost a third of which with depression. The data came from a variety of sources, including the personal genomics company 23andMe. Individuals either self-diagnosed depression symptoms or had a clinical diagnosis when they had their genetic information collected. These individuals were then compared with the control samples and 44 loci were identified as associated with depression. To put it another way, individuals with certain genetic variants, or alleles, in these regions are statistically more likely to have depression.
To get these large sample sizes, we used diverse studies. From recurrent, severe, clinically-ascertained depression to self-report depression (yes/no) from @23andMe. All sources contributed, all showed similar signal for genetics of depression (4/8) pic.twitter.com/pLUoa4X39a
— Cathryn Lewis (@cathrynlewis) 26 April 2018
The results of this study raises the hope that future research will be able to look at these loci in more detail and that further information on the genetic components of depression can be inferred, and ultimately more personalised and effective treatment given.
Another interesting finding from this research is that the same genes associated with depression also show significant relationships with other psychiatric disorders, such as anxiety and autism spectrum disorders. Other associations were also found, with less education and higher body mass linked to these same genetic regions.
IMPC data
A total of 155 genes were identified from the 44 loci mentioned in this study. Of these genes, 46 knockout mice have been produced through the IMPC. From these 46 IMPC genes 31 have phenotype data available. 6 show abnormal behaviour related phenotypes, including hyperactivity, hypoactivity, abnormal sleep behaviour and abnormal behavioural response to light. IMPC data was not included in the study but this does point to some interesting associations that may be relevant for future research.
| Gene | All phenotypes |
| Emilin3 | Hyperactivity, abnormal behavior |
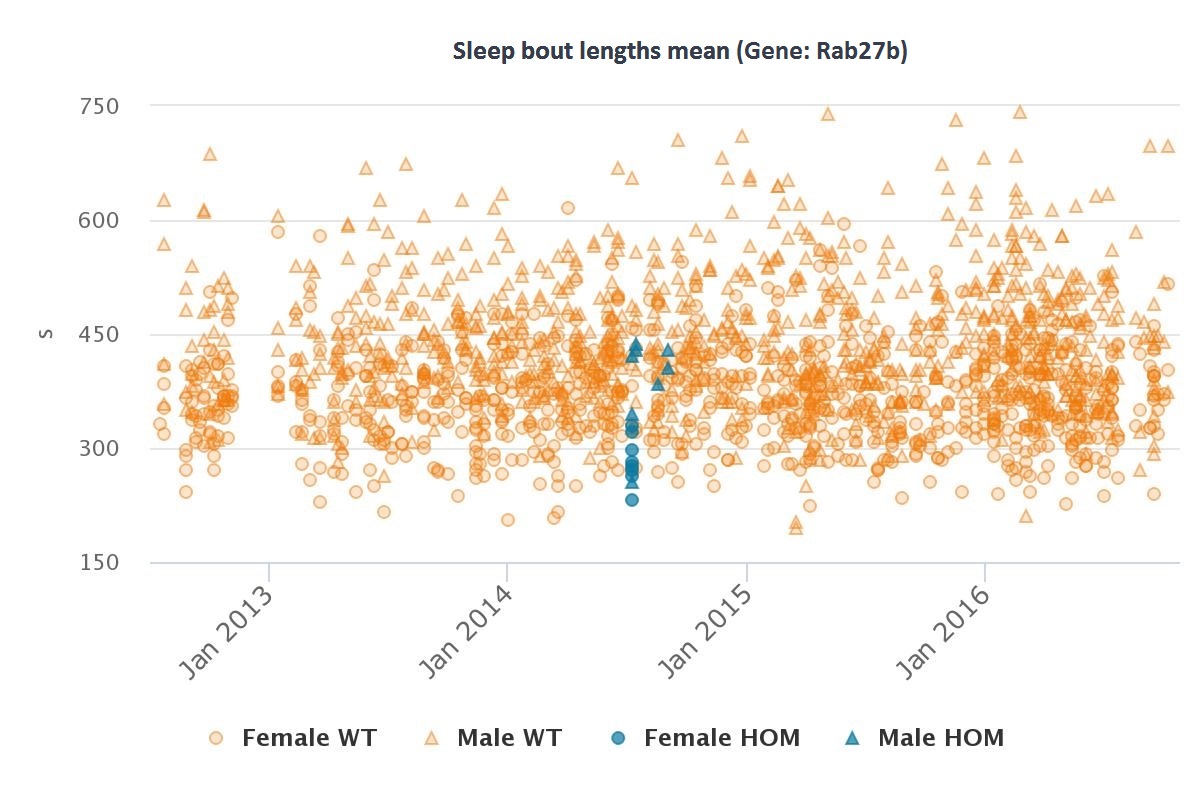
| Rab27b | Abnormal sleep behavior |
| Kcnb1 | Increased mean corpuscular volume, hyperactivity, abnormal gait, abnormal locomotor behavior, abnormal behavior |
| Fads1 | Hypoactivity, improved glucose tolerance, abnormal seminal vesicle morphology, abnormal testis morphology, small testis |
| Pax5 | Increased circulating triglyceride level, hyperactivity, decreased leukocyte cell number, impaired glucose tolerance, decreased threshold for auditory brainstem response |
| Zscan12 | Abnormal behavioral response to light, abnormal bone structure |
Terry Meehan, a member of IMPC and not involved in this study, said: “Human GWAS studies identify small regions of the genome that are associated with complex traits like depression. While small, these regions may encompass dozens of genes. It is often difficult to tell which genes within these regions are contributing to the trait and which are passengers. IMPC phenotype data is now of sufficiently broad scale that we can help clinical geneticists find those causative genes”.